a National Influenza Center, National Institute of Hygiene and Epidemiology, Hanoi, Vietnam.
b Military Hospital 108, Hanoi, Vietnam.
Correspondence to Hoang Vu Mai-Phuong (e-mail: lom9@nihe.org.vn or hoangmp9906@gmail.com).
To cite this article:
Hoang Vu MP et al. Oseltamivir resistance among influenza viruses: surveillance in northern Viet Nam, 2009–2012. Western Pacific Surveillance and Response Journal, 2013, 4(2):25–29. doi:10.5365/wpsar.2013.4.1.010
Introduction: Antiviral resistance has been reported in seasonal influenza A viruses and avian influenza A(H5N1) viruses in Viet Nam, raising concerns about the efficacy of treatment.
Methods: We analysed specimens from two sources during the period 2009–2012: influenza-positive samples from influenza-like illness patients at sentinel clinics in northern Viet Nam and isolates from patients with confirmed A(H5N1) infections. Pyrosequencing was used to detect mutations: H275Y [for A(H1N1) and A(H5N1)], E119V [for A(H3N2)] and I117V [for A(H5N1)]. A neuraminidase inhibition assay was used to determine the Inhibitory Concentration 50 (IC50) values for all influenza A and B isolates.
Results: There were 341 influenza A positive samples identified; influenza A(H1N1)pdm09 was identified most frequently (n = 215). In 2009, oseltamivir resistance was observed in 100% (19 of 19) of seasonal A(H1N1) isolates and 1.4% (3/215) of A(H1N1)pdm09 isolates. This H275Y mutation was not found in influenza subtypes A(H5N1) or A(H3N2) isolates.
Discussion: In Viet Nam, seasonal and A(H5N1) influenza vaccines are not currently available; thus, effective treatment is required. The presence of oseltamivir-resistant viruses is therefore a concern. Active surveillance for oseltamivir resistance among influenza viruses circulating in Viet Nam should be continued.
Influenza infection causes annual epidemics throughout the world. There are two common types of influenza viruses that cause human infection – influenza A and influenza B. Influenza A viruses caused several influenza pandemics in the 20th century, and a pandemic caused by the influenza A(H1N1)pdm09 virus occurred in 2009.1 National influenza surveillance was initiated in Viet Nam in 2006, and the data collected so far have shown that influenza viruses circulate year-round with similar peaks and subtypes observed across all surveillance regions.2 Between 2003 and 2012, 123 human cases of A(H5N1) infection were confirmed from 40 of the 63 provinces in Viet Nam, with 81 cases (66%) from northern Viet Nam.3 Although influenza vaccines that protect against A(H1N1)pdm09 or influenza A(H5N1) are being developed in Viet Nam, they are currently only available through private market purchase.
The neuraminidase inhibitors oseltamivir and zanamivir are the primary antiviral agents recommended for the treatment of influenza infections,4,5 yet antiviral resistance to influenza A viruses is increasingly being reported.6,7 Oseltamivir is currently recommended as the first-line option by the Viet Nam Ministry of Health for treating suspected infections of A(H5N1) and A(H1N1)pdm09. The emergence of oseltamivir resistance of clinical isolates of influenza A virus has been associated with substitution at residue V116, I117, E119, Q136, K150, D151, D199, I223, H275 and N295 in the neuraminidase active site.8 For influenza B there have been two main substitutions: residues R152 and D198.8,9 In Viet Nam, oseltamivir-resistant strains harboring mutations at positions I117V, H275Y and N295S were reported for A(H5N1) in 2005,6 A(H1N1) in 200710 and A(H1N1)pdm09 in 2009.7,11 The limitations of other antiviral drugs, as well as the risk of oseltamivir resistance, have raised concerns about the efficacy of oseltamivir for influenza infection treatment. We report here on a pilot study for the establishment of a routine antiviral resistance surveillance programme in northern Viet Nam.
As an initial step in establishing a surveillance programme for antiviral resistance in northern Viet Nam, genetic analysis was conducted for both clinical specimens and isolates collected through sentinel sites and isolates of influenza A(H5N1). Neuraminidase activity was measured using a phenotypic method for viral isolates of influenza A and B. Pyrosequencing assays were then applied to detect the common mutations related to reducing susceptibility or resistance of influenza A viruses to oseltamivir – I117V, E119V and H275Y. Data from the National Influenza Surveillance System in Viet Nam were also analysed for the period 2009–2012.
Throat swabs were collected as part of the sentinel surveillance system for influenza-like illness in northern Viet Nam between 2009 and 2012 and were screened using standard methodology (conventional reverse transcriptase polymerase chain reaction, RT–PCR) at the National Influenza Center of the National Institute of Hygiene and Epidemiology in Hanoi.2 Influenza isolates were then cultured from throat swabs positive for influenza A and B.2,6 In addition, isolates from throat swabs, pharyngeal swabs or tracheal swabs collected from A(H5N1)-infected patients admitted to intensive care units of general hospitals in northern Viet Nam between 2009 and 2012 were obtained.
Madin-Darby canine kidney (MDCK) cells, obtained from the American Type Culture Collection, were used to culture viruses. Swabs positive for influenza A(H5N1) by RT–PCR were inoculated biosafety level III culture facilities. Viruses were harvested and stored at −80 ˚C for further analysis. All influenza isolates with a minimum of eight haemagglutination units by haemagglutination inhibition assay were selected for neuraminidase inhibition assay.6,8,10,11
Pyrosequencing assays were used to further characterize all RT–PCR influenza-positive samples (n = 341) and influenza isolates (n = 67). Viral RNA was extracted directly from clinical specimens or from supernatants of isolates propagated in MDCK cells by using a viral RNA extraction kit (Qiagen, USA) according to the manufacturer’s instructions. RT–PCR amplification was conducted using the Qiagen PyroMark PCR kit with specific primer sets for A(H1N1)pdm09, seasonal A(H1N1), A(H3N2) and A(H5N1).12,13 Three sets of RT–PCR primers were used to generate corresponding amplicons of the neuraminidase gene segment covering the sequences encoding the target residues 117, 119 and 275 according to procedures described previously.14
The pyrosequencing reactions and data analysis were performed using a PyroMark Q24 ID Platform (Qiagen, USA). Briefly, biotinylated PCR products were washed through a series of buffers, and single-stranded DNA was generated and used as a template for hybridization to residue-specific sequencing primers, which were used at a concentration of 100 µM.11,15
Oseltamivir carboxylate (GS4071) and its active form (GS4104) were provided by Roche Laboratories, Inc, Basel, Switzerland. The reference influenza viruses were provided by the World Health Organization (WHO) Collaborating Centre for Reference and Research on Influenza, Melbourne, Australia. Influenza A(H5N1) isolates were inactivated by 1% formalin for 24 hours before the NAI assay. The NAI assay was performed according to procedures described previously.8
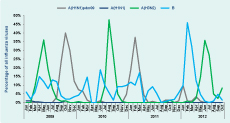
During the period 2009–2012, there was year-round circulation of seasonal influenza viruses with frequent co-circulation of influenza A and influenza B (Figure 1). A total of 341 influenza A positive samples were identified by RT–PCR (Table 1). Of these, influenza A(H1N1)pdm09 was identified most frequently (n = 215) throughout the whole study period; 100 were A(H3N2) and seven were A(H5N1). Seasonal A(H1N1) was isolated only in 2009 (n = 19).

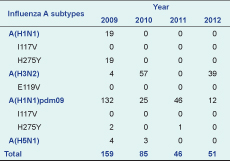
Oseltamivir resistance, as determined by detection of H275Y in the neuraminidase gene, was observed in 100% (19 of 19) of seasonal A(H1N1) isolates in 2009 and was identified in 1.4% (3/215) of A(H1N1)pdm09 isolates collected in 2009. This H275Y mutation was not found in influenza subtypes A(H5N1) or A(H3N2) isolates. I117V was not observed in any of the isolates of subtypes A(H1N1), A(H5N1) or A(H1N1)pdm09; I119V also was not found in any A(H3N2) isolates (Table 1).
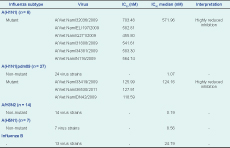
There were 67 isolates that underwent NAI assay; six seasonal A(H1N1), 14 A(H3N2), seven A(H5N1), 27 A(H1N1)pdm09 and 13 influenza B. All of the seasonal A(H1N1) isolates (n = 6) had Inhibitory Concentration 50 (IC50) values that ranged from 541.61 to 703.48 nM higher than the reference virus (A/Mississippi/3/2001, oseltamivir-resistant) and reached 1000-fold higher than the reference wild-type virus (A/Mississippi/3/2001, oseltamivir-sensitive). Among the 27 viruses of A(H1N1)pdm09, IC50 values ranged from 118.59 to 127.91 nM and reached 250-fold higher than the reference wild type virus. The IC50 values obtained from non-mutant viruses [A(H1N1)pdm09, A(H3N2), A(H5N1) or influenza B] had median IC50 ranging from 1.07 nM [A(H1N1)pdm09] to 8.56 nM [A(H5N1)] and 24.79 nM (B), i.e. sensitive to oseltamivir (Table 2).
IC50 – Inhibitory Concentration 50
Our study of oseltamivir-resistant influenza viruses in northern Viet Nam shows that seasonal A(H1N1) isolates circulating in 2009 were oseltamivir-resistant by virtue of having the H275Y mutation and IC50 values indicative of resistance. This finding is consistent with reports from Japan, the United States of America and Europe of high rates of resistance (100%) during the 2008 to 2009 season.12,13,15 WHO also reported the spread of resistant A(H1N1) strains worldwide during that time.
The A(H1N1)pdm09 virus was first detected in June 2009 in Viet Nam and was the predominant virus during the 2009 and 2011 influenza seasons. However, in this study, the oseltamivir-resistent mutation (H275Y) was identified in only two specimens in 2009 and one in 2011, a rate of 1.5% in 2009. A separate cluster of seven cases of oseltamivir-resistant A(H1N1)pdm09 was also reported from Viet Nam in July 2009.5 The rate from this study is consistent with that collected through the WHO Global Influenza Surveillance and Response System of 1.7% frequency of resistance of A(H1N1)pdm09 in the first two years of the pandemic (2009–2010).16,17 Similar data reported elsewhere ranged from 0.5% in the United States of America to 0.8% in the United Kingdom and 1.1% in the Asia-Pacific.12,14,18–20
In our study, A(H5N1), A(H3N2) and B viruses were determined to be oseltamivir-sensitive by genotypic and phenotypic testing. These results are reassuring for future treatment of A(H5N1) infections in Viet Nam with oseltamivir, as the A(H5N1) influenza vaccine is not available. However, an oseltamivir-resistant A(H5N1) virus was reported in human isolates in 2005, and the emergence of mutations associated with reducing susceptibility (I117V) to oseltamivir was also determined among A(H5N1) isolates from both human and poultry in 2009–2010. Thus, continuing oseltamivir-resistance surveillance is critical for public health as oseltamivir is the most widely used antiviral medication for H5N1 infection.16
This study is subject to several limitations. The main limitation is the sample size of both biological and viral isolates, as we experienced difficulty in growing influenza A(H3N2) and A(H1N1)pdm09 and the quality of samples caused a reduced number of viral isolates to be tested. Also, it should be noted that phenotypic data (and sequencing) can only indicate that a virus is resistant: a direct relationship between IC50 values and actual clinical resistance is yet to be proven. Our data represent northern Viet Nam only and might not provide an accurate picture of the prevalence of oseltamivir-resistant viruses in the whole country. The data collected from the national influenza surveillance system did not report periodic antiviral use, and therefore it is difficult to assume that any resistance found in our study was due to transmitted resistance. Active surveillance in the future should be expanded to include data on oseltamivir use in hospitals and private clinics.
In conclusion, phenotypic and sequencing data indicated that oseltamivir resistance was present in seasonal A(H1N1) and A(H1N1)pdm09 subtypes in Viet Nam during the period 2009–2012. An increase of antiviral-resistant influenza viruses might occur in Viet Nam in the future. Enhancing active surveillance by expanding this pilot study to different regions, monitoring the use of oseltamivir, analysing more specimens and reviewing more epidemiological information is recommended for Viet Nam.
None of declared.
This study was supported by a grant from the Korea Center for Diseases Control.
We are grateful the National Institute of Hygiene and Epidemiology for their continued support for our works, we would like to thank technical support from WHO Viet Nam and WHO Collaborating Center, Melbourne, Australia.