a Swiss Tropical and Public Health Institute, Basel, Switzerland.
b University of Basel, Basel, Switzerland.
c Papua New Guinea Institute of Medical Research, Goroka and Madang, Papua New Guinea.
d Queensland Mycobacterium Reference Laboratory, Pathology Queensland, Brisbane, Australia.
Correspondence to Hans-Peter Beck (email: Hans-Peter.Beck@unibas.ch).
To cite this article:
Ley S et al. Non-tuberculous mycobacteria: baseline data from three sites in Papua New Guinea, 2010–2012. Western Pacific Surveillance and Response Journal, 2015, 6(4);24–29. doi:10.5365/wpsar.2015.6.2.004
Objective: To determine the proportion of non-tuberculous mycobacteria (NTM) in samples of pulmonary tuberculosis (TB) cases from Papua New Guinea who were diagnosed using acid-fast microscopy.
Methods: As part of a case detection study for TB, conducted in three provincial hospitals in Papua New Guinea, sputum samples of suspected tuberculous cases aged 15 years or older were collected from November 2010 to July 2012. Mycobacterial species isolated from sputum and grown in culture were examined to distinguish between NTM and the Mycobacterium tuberculosis complex (MTBC).
Results: NTM were detected in 4% (9/225) of sputum samples grown in culture. Five (2.2%) of them were identified as NTM only and four (1.8%) were identified as mixed cultures containing both MTBC and NTM. Four different NTM species were identified; M. fortuitum, M. intracellulare, M. terrae and M. avium.
Discussion: This is the first report from Papua New Guinea identifying NTM in three different locations. As NTM cannot be distinguished from M. tuberculosis through smear microscopy, the presence of NTM can lead to a false-positive diagnosis of tuberculosis. The prevalence of NTM should be determined and a diagnostic algorithm developed to confirm acid-fast bacilli in a smear as M. tuberculosis.
Apart from the Mycobacterium tuberculosis complex (MTBC), the genus Mycobacterium includes over 120 species of non-tuberculous mycobacteria (NTM).1 NTM can be found in the environment, including water and soil, which is the suspected source of occasional infection of humans. Asymptomatic colonization as well as symptomatic disease can be caused by NTM,2 including, among others, chronic pulmonary disease with symptoms similar to tuberculosis (TB) such as chronic cough (with or without sputum production), chest pain and weight loss.3,4 Different NTM have been associated with different disease presentations. The M. avium complex (including M. avium and M. intracellulare) is most often associated with pulmonary infection. M. fortuitum has been associated with pulmonary infection but more often affects the skin, soft tissue or bones. Immunocompromised cases (e.g. human immunodeficiency virus [HIV] positive cases) are susceptible to NTM infection, particularly disseminated M. avium disease.2 However, immunocompetent cases with no predisposing conditions can also be affected.5–8
Standard first-line anti-TB treatment drugs are less effective against NTM compared to M. tuberculosis (Mtb),2,9 and no single regimen for NTM exists to date. Depending on the NTM species, recommendations for treatment regimens include treatment with antibiotics and sometimes even surgical removal of infected tissue.2,10 The M. avium complex is treated with combination therapy consisting of Clarithromycin, Rifampicin and Ethambutol and should be continued for one year.11 While the regimen includes Rifampicin and Ethambutol, two of the standard first-line anti-TB drugs, the length of the TB regimen is not sufficient to address M. avium complex infections. Additionally, Isoniazid (apart from Rifampicin the most potent first-line anti-TB drug) has only a limited effect on M. avium,9 and relapses are common.2
Little data are available on the prevalence of NTM infections in TB high-burden countries, but the incidence can nevertheless be substantial.12 High TB-burden countries also tend to be resource-poor countries, and the diagnosis of pulmonary TB is based on the microscopic detection of acid-fast bacilli (AFB) in sputum samples. Smear microscopy cannot distinguish between NTM and Mtb. Mixed infections as well as false-positive TB diagnosis cannot be ruled out. Many diagnostic assays are not optimized to detect different NTM species; if NTM are present in conjunction with Mtb, the former might remain undetected or cause failure of drug susceptibility testing (DST).13–15 Exposure to NTM has been suggested to impact on the efficacy of the Bacille Calmette-Guérin vaccine16 and to exhibit cross-reactivity to the tuberculin skin test (TST), leading to increased difficulties in interpreting TST-positive results and evaluating the protection through the only available vaccine against TB.17,18
Very little information is available on NTM in Papua New Guinea. Data from a leprosy trial conducted in Karimui (Eastern Highlands Province) in the 1960s19,20 as well as a TST sensitivity study conducted in the Marawaka area of the Eastern Highlands of Papua New Guinea21 found no evidence for environmental mycobacteria being present in this area. Therefore it was important to investigate the presence of NTM in sputum samples collected in Papua New Guinea. Here we describe the NTM detected and provide baseline information on these bacteria in Papua New Guinea.
As part of a case detection study for TB, conducted between November 2010 and July 2012 in selected provincial hospitals in Papua New Guinea, sputum samples of suspected TB cases aged 15 years or older were collected for laboratory testing. The sampling procedure has been described previously.22
Upon diagnosis of TB through AFB Ziehl-Neelson (ZN) microscopy or chest X-ray, sputum samples were decontaminated following Petroff’s method;23 inoculated into BD Bactec® Mycobacterial Growth Indicator Tube (MGIT) media (Becton, Dickinson and Co., Franklin Lakes, New Jersey, USA); and subsequently sent to the Queensland Mycobacterium Reference Laboratory in Brisbane, Australia for culture. The samples were incubated in the MGIT until they became culture positive (i.e. growth could be detected). A repeat ZN smear was prepared on all culture-positive isolates to confirm the presence of acid-fast organisms. A rapid immuno-chromatographic identification test (SD BIOLINE/BD TB Ag MPT64 Rapid, Standard Diagnostics, Giheung-gu, Republic of Korea) was used to confirm the AFB as MTBC.
When the rapid test was negative or the microscopic morphology did not suggest the AFB were MTBC, further molecular analysis was conducted to identify the isolate as NTM or MTBC. In brief, DNA was extracted using crude boil method at 95 °C for 30 minutes, followed by sonication for 15 minutes. The extracted DNA was then used as a template for polymerase chain reaction (PCR) amplification either according to the GenoType® Mycobacterium Common Mycobacteria line probe kit (Hain LifeSciences, Nehren, Germany) according to the manufacturer’s protocol for the GenoType 16S rRNA (Forward primer 5′ AGAGTTGGATCCTGGCTCAG; Reverse primer 5′ CCTACGAGCTCTTTACG). The amplified product was purified using 4ul EXOSAP-IT (Affymetrix, San Diego, California, USA) and 10ul of primary amplification product (37 °C 15 minutes, 80 °C 15 minutes, 40 °C soak). A repeat gel was run using Invitrogen Bufferless Gel system (ThermoFisher Scientific, Waltham, Massachusetts, USA). The sequencing reaction was performed using the Big Dye Terminator method on ABI3130 sequencer (Distribio, Dudelange, Luxembourg), and the resulting sequences were analysed by comparing them to the National Center for Biotechnology Information Genbank database. In case cultures were identified as MTBC, DST was subsequently performed by the proportion method,24 as described previously.25 However, if a culture turned out to be NTM, no DST was performed.
Demographic and clinical symptoms of the cases were also collected for analysis. Statistical analysis was carried out with Stata 12.1 (Stata-Corp, College Station, Texas, USA). Excel was used for basic calculations. Due to a small sample size, no statistical analysis for the NTM population was performed.
Ethical approval for this study was granted by the Papua New Guinea Institute of Medical Research Institutional Review Board (IRB No. 0913) and the Papua New Guinea Medical Research Advisory Council (MRAC No. 10.02). The Ethik-Kommission beider Basel has been informed and had approved the study. Written informed consent was obtained from all study participants.
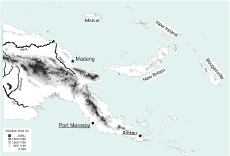
A total of 396 sputum samples were collected in three provincial hospitals in Papua New Guinea (Figure 1). Of the collected samples, 335 were sent to Australia for culture and 225 samples grew in culture. NTM were detected in 4% (9/225) of those samples. Five (2.2%) samples contained a NTM only, consisting of three isolates of M. fortuitum, one isolate of M. terrae and one isolate of M. intracellulare. Four (1.8%) isolates were identified as mixed cultures containing both bacteria of the MTBC and NTM. These included three cultures of MTBC and M. avium and one culture of MTBC and M. intracellulare (Table 1).

Note: Map designed by authors using MapInfo Professional 7.0.
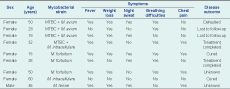
MTBC, Mycobacterium tuberculosis complex; and NTM, non-tuberculous mycobacteria.
All but one of the NTM infections were detected in females. All the cases with either a mixed infection or a NTM infection only had reported productive coughs for at least two weeks. All the cases with a mixed infection of MTBC and NTM additionally reported weight loss and at least one other symptom, including breathing difficulties (n = 3), chest pain (n = 3) fever and night sweats (n = 2). Among the five cases with an NTM infection only, four cases reported shortness of breath and fever. Three of those cases experienced weight loss and either chest pain or night sweats, or both. The case infected with M. intracellulare reported no other symptoms except for productive cough and headache. None of the cases had reported any previous TB episode (Table 1).
To our knowledge this is the first study describing the presence of NTM in Papua New Guinea. In five (2.2%) of the 225 cases, the isolate was identified as a NTM. Without culture results from at least one more follow-up sample, this may indicate several false-positive TB cases. General symptoms caused by NTM infections cannot be distinguished from symptoms observed in TB cases, and the appearances of the bacteria cannot be differentiated when examined by AFB ZN light microscopy.
It is interesting that in our case cohort all but one NTM isolates were found in females; the only isolate identified in a male was M. terrae. There are some NTM species which were more commonly isolated from females.2,7,26 Another study showed an increased prevalence of funnel chest (pectus excavatum) and abnormal narrowing of the thoracic dimension in female cases infected with NTM of the M. avium complex not seen in males.26 Also, the so-called Lady Windermere syndrome, a specific pulmonary disorder caused by bacteria of the M. avium complex, was only found in women.27
There are only a few reports on NTM from TB-endemic countries,3 and it is generally difficult to compare our findings with studies from other countries. In a recently published study from Nigeria, for example, 15% of culture-grown mycobacteria isolated from presumptively diagnosed pulmonary TB cases were NTM.28 Compared to that study, a ratio of 2.2% in our study is relatively low. However, culture criteria of these two studies differed. Whereas in our study only smear-positive samples were cultured. A 2013 study also included smear-negative samples, which turned out to be more strongly associated with NTM infections than smear-positive samples.28 It is likely that limiting culture to smear-positive isolates in our study has reduced the chances of detecting NTM in sputum. However, culturing smear-positive samples only is in accordance with the protocols of the National TB Programme of Papua New Guinea and a result of logistic challenges arising from the lack of an in-country culture facility.
Our study population was furthermore limited to suspected pulmonary TB cases aged 15 years or above from three sites within Papua New Guinea, and it is unclear whether inferences can be made to the rest of Papua New Guinea. Nevertheless, compared to the few studies conducted in Papua New Guinea in the 1960s and 1980s,19–21 where tuberculin skin testing did not provide evidence for NTM, our results highlight the existence of NTM in the community and the potential impact on TB diagnosis in the country. While the possibility remains that the presence of NTM in sputum specimens is due to colonization with these environmental organisms, they can also lead to false-positive TB diagnosis when AFB smear microscopy is used alone. The standard anti-TB treatment is not ideal for NTM, as different antibiotics than the ones used against TB are required to treat NTM,2,10 leading to an additional burden for the case as well as the National TB Programme. With an increasing burden of HIV/AIDS, NTM may also become an increasing source of disease, requiring different approaches for case management and treatment.
In Papua New Guinea, the diagnosis of multidrug-resistant (MDR) TB was for a long time based on the observation of repeated treatment failure despite compliance with treatment.29 Since 2012, TB drug resistance surveillance based on Xpert® MTB/RIF assay (Cepheid, Sunnyvale, California, USA) has started in a few major cities.30 However, it probably remains difficult for many health facilities to obtain a culture/DST-confirmed diagnosis of MDR-TB. If the actual cause of treatment failure is not drug resistance, but an NTM infection, this would have a major impact on individual case management, especially if the symptoms of the disease are similar to those of MDR-TB. This has been shown in a study from India, where 17.6% of the suspected MDR pulmonary TB cases were actually NTM infections.3 An additional challenge to the laboratory is the presence of mixed infections of NTM and MTBC; reliable DST for MTBC may be difficult if the strain cannot be isolated in pure culture, leading to false positivity including incorrect designation of MDR-TB and extensively drug-resistant TB.
As our sample size of detected NTM is small, further studies are required to obtain significant data to establish a valid diagnostic algorithm and treatment guidelines for pulmonary diseases caused by NTM. However, no NTM identification is yet performed in the framework of the National TB Programme in Papua New Guinea, and to date, no biosafety level 3 laboratory required for culturing mycobacteria is available in the country. Samples from cases suspected of having MDR-TB are shipped to a mycobacterium reference laboratory in Australia for culture. In-country mycobacterial culture would distinguish TB from NTM infections much more rapidly and at the same time improve the detection of drug-resistant TB.
It is recommended that NTM infection surveillance could be added to the TB drug resistance surveillance of the National TB Programme.30 Data from NTM surveillance would determine NTM’s role in pulmonary disease in Papua New Guinea and would inform health authorities to target interventions and response in the future. This would relieve both cases and the health system. As Xpert® MTB/RIF assay is not detecting NTM, smear-positive but Xpert® MTB/RIF-negative results could be used as an indicator for NTM infection and as a basis for further investigation. Until culture becomes available within the country, PCR-based assays amplifying the internal transcribed spacer region of 16–23S rRNA could be implemented at the country’s Central Public Health Laboratory to distinguish NTM from MTBC directly from clinical samples.31
None declared.
This research was conducted in the framework of a TB passive case detection study funded by the Stanley Thomas Johnson Foundation and the Medicor Foundation Liechtenstein.
We thank all study participants who agreed to having their samples collected and analysed. We further thank the health authorities of all three study provinces for approval and general support. We are indebted to the Papua New Guinea Institute of Medical Research study teams and the hospital staff in all three sites for sample collection, diagnosis, access to infrastructure and general support. Also, the contribution of the staff of the Queensland Mycobacterium Reference Laboratory is gratefully acknowledged.