a Charles Sturt University, New South Wales, Australia.
b University of Queensland, Brisbane, Australia.
Correspondence to Thiru Vanniasinkam (e-mail: tvanniasinkam@csu.edu.au).
To cite this article:
Detection of Campylobacter in human faecal samples in Fiji. Western Pacific Surveillance and Response Journal, 2014, 5(4):30–33. doi:10.5365/wpsar.2014.2.007
Introduction: Data on campylobacteriosis in developed countries are well documented; in contrast, few studies on campylobacteriosis have been conducted in developing countries. This study was undertaken to test for Campylobacter in human faecal samples sent to the two major pathology laboratories in Fiji.
Methods: A total of 408 diarrhoeal faecal samples were collected from the two major hospital pathology laboratories in Central Fiji (Suva) and Western Fiji (Lautoka) between December 2012 and February 2013 and from June to July 2013. Samples were analysed for the presence of Campylobacter using polymerase chain reaction (PCR) based methods.
Results: Campylobacter was detected in 241/408 (59.1%) of samples tested using PCR. Samples from children aged less than five accounted for 21.6% of positive cases.
Discussion: Campylobacter was detected in 59.1% of diarrhoeal samples collected from the two main laboratories in Fiji. A high proportion of children under five years with Campylobacter has been reported in other countries and could be due to parents being more likely to seek medical attention. Further studies are required to confirm the species of Campylobacter that are predominantly associated with gastroenteritis in Fiji.
Campylobacter spp. have been recognized as gastrointestinal pathogens in both developed and developing countries and are ubiquitous in food animals such as poultry, cattle, pigs, sheep, ostriches and shellfish and in pets such as cats and dogs.1 Patients with campylobacteriosis present with symptoms similar to those seen in other enteric infections, and while symptoms are usually self-limiting, in severe cases symptoms may last for 5 to 7 days.2
Data on campylobacteriosis not exist in many developing countries due to the lack of surveillance programmes for Campylobacter infections. It has been reported that Campylobacter infections in developing countries are more frequently reported in children under five years of age and often regarded as a paediatric disease.3 Based upon current studies, the rates of campylobacteriosis in the general population are an estimated 90 cases per 100 000 population in both developing and developed countries.3
Diarrhoeal disease is notifiable in Fiji; approximately 22 753 diarrhoeal cases and 281 cases of diarrhoea with blood were reported by the Ministry of Health in Fiji in 2011.4 Salmonella typhi infections, classified as enteric fever, were detected in 404 of these cases;4 however, no other pathogen-specific data were reported. This is despite studies showing widespread prevalence of gastrointestinal disease in developing countries.5 The public health infrastructure is variable in the Pacific region. Many countries are geographically isolated and have limited economic resources;6 therefore, specific communicable diseases including foodborne disease are neither notifiable nor monitored through laboratory-based surveillance systems.
In Fiji, when stool samples are collected, most pathology laboratories routinely screen for parasites, viruses and bacterial pathogens such as Salmonella and Shigella but not for Campylobacter.
This study was undertaken to test for Campylobacter in stool samples sent to the two major pathology laboratories in Fiji using polymerase chain reaction (PCR) methods.
This study was approved by the Charles Sturt University Human Ethics Committee and the National Research Ethics Committee, Ministry of Health, Fiji.
A total of 408 human faecal samples (stool samples) were collected from the Central and Western hospitals’ pathology laboratories in Fiji from mid-December 2012 to the end of February 2013 and from June to July 2013. The samples were approximately equally divided between the two sites (Central [Suva] n = 208, Western [Lautoka] n = 200). Information on age and gender were extracted manually from the stool register held by the laboratories; due to time and access constraints, it was not possible to obtain this information for all samples.
Samples were collected in sterile containers, placed on ice and transported to the Fiji National University laboratory for PCR analysis. Samples were only made available to this study after completion of routine testing by the microbiology department in both laboratories: four to six hours after the sample was received in the laboratory in Suva and eight to 10 hours in Lautoka.
Total DNA was extracted from the faecal samples using the Norgen DNA Stool Mini Kit (Norgen Biotek Corporation, Thorold, ON, Canada). An aliquot of the stool sample (200µL) was collected in 2ml Eppendorf tubes and heated at 70 °C for 10 minutes. The DNA was extracted using the manufacturer’s protocol. The final elution volume of DNA was 100µl. Extracted DNA samples were stored at –20 °C before PCR analysis.
PCR amplification was performed using previously described primers of a fragment specific for the 16SrRNA gene, C412F,5′-GGATACACTTTTCGGAGC-3′ and C1288R,5′-CATTGTAGCACGTGTGTC-3′.7 Each PCR reaction (25µl) contained GoTaq Green Master Mix 2x (Promega, Madison, WI, USA) that was used as the reaction buffer with 400µM dATP, 400µM dGTP, 400µM dCTP, 3mM MgCl2 and 50 µM of each primer. DNA template (1µl) was used and subjected to 35 cycles of amplification in a thermal cycler (Eppendorf) with the following slightly modified conditions: denaturation at 94 °C for 5 minutes, annealing temperature for 16SrRNA, 58 °C for 30 seconds, extension for 72 °C for 1 minute and further extension at 72 °C for 5 minutes.7 The PCR amplicons were analysed by 1% agarose gel electrophoresis stained with ethidium bromide and visualized. All PCR assays were set up using DNA from Campylobacter jejuni NCTC 11351 as the positive control. No bacterial DNA was added to the negative control. Results were discarded if the negative control was positive. Species level identification was not undertaken on the Campylobacter-positive samples.
Of the 408 (Suva, n = 208; Lautoka, n = 200) stool samples analysed by PCR, 241 (59.1%) were positive for Campylobacter. A significantly higher proportion of PCR-positive samples were detected from the Central area (Suva, n = 141; 67.8%) than from the Western area (Lautoka, n = 100; 50%; P < 0.008).

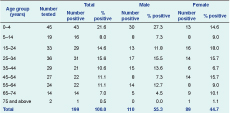
* Age and gender data were not available for 42 positive samples.
Valid information was available on patient age for 229 samples and sex for 236 samples (132 males and 104 females). There was no significant difference regarding sex or age group distribution of cases between the Central and Western regions (P = 0.982 and P = 0.357, respectively). Although there were more PCR-confirmed Campylobacter infections in males compared to females (55.3% compared with 44.7%), this was not statistically significant (P = 0.125). The highest proportion of positive cases was observed in the 0–4 year age group at 21.6%, followed by a second peak (15%) in the 15–34 years age group (Table 1).
This study reports the detection of Campylobacter in faecal samples collected from patients with diarrhoea in Fiji from mid-December 2012 to February 2013 and in June and July 2013. In the study population, Campylobacter was detected in the majority (59%) of clinical samples. This finding is similar to a study undertaken in United States military personnel training in Thailand.8
The age distribution of PCR-confirmed cases of campylobacteriosis in this study was similar to the findings of other studies in developing countries, with a high proportion of cases in children aged less than five years.5 One such example is a study carried out in Malawi where 21% of children under five years of age with diarrhoea had Campylobacter infections, and 14% of Campylobacter infections were detected in asymptomatic children.9 Children may be more likely to be taken to hospitals by their parents when exhibiting symptoms of gastrointestinal disease, and consequently, faecal samples from these patients are frequently submitted to pathology laboratories for testing.
The sources of Campylobacter spp. infections in Fiji are not known. Poor food handling techniques and hygiene have been reported as the cause of diarrhoeal disease, and epidemiological studies also report cases of campylobacteriosis occurring through the consumption of raw milk9 and undercooked or contaminated poultry meat.10 All of these are possible risk factors in Fiji.
The diagnosis of Campylobacter infection is typically performed using culture techniques and in some cases PCR.10 An important advantage of using PCR is that it can detect dead Campylobacter spp. in clinical samples, which may be important when poorly stored samples are analysed. Currently, in Fiji, pathology laboratories do not routinely test diarrhoeal samples for the presence of Campylobacter. To the best of the authors’ knowledge, this is the first study to investigate the prevalence of Campylobacter spp. in diarrhoeal samples in Fiji.
The high rate of Campylobacter detected in this study requires further investigation; future studies should include healthy participants to determine the rate of Campylobacter carriage in the Fiji population. It would also be useful to determine the species of Campylobacter isolated in this population. Additionally, it would be valuable to determine if pathogens other than Campylobacter were present in the Campylobacter-positive samples from individuals with diarrhoea. Coinfection with Salmonella typhi or other pathogens were not investigated in this study.
In conclusion, the results of this study demonstrate that Campylobacter infections are common in Fiji, similar to many other developing and developed countries.
This study was funded by Charles Sturt University, COMPACT Scholarship and College of Medicine, Nursing and Health Sciences, Fiji National University for the doctoral project of Aruna Devi.
None declared.
The authors thank the Ministry of Health, Fiji for the approval of this study and the College of Medicine, Fiji National University for allowing us to use their laboratory facilities. The authors also thank the staff of the microbiology department, Suva and Lautoka pathology laboratories for providing the samples for the study.