
a Victorian Infectious Diseases Reference Laboratory, North Melbourne, Victoria, Australia.
b Nha Trang Pasteur Institute, Nha Trang, Viet Nam.
c Ninh Hoa General Hospital, Khanh Hoa, Viet Nam.
Correspondence to Thomas Tran (e-mail: thomas.tran@mh.org.au).
To cite this article:
Tran T et al. Respiratory virus laboratory pandemic planning and surveillance in central Viet Nam, 2008–2010. Western Pacific Surveillance and Response Journal, 2012, 3(3):49-56. doi: 10.5365/wpsar.2012.3.2.011
Introduction: Laboratory capacity is needed in central Viet Nam to provide early warning to public health authorities of respiratory outbreaks of importance to human health, for example the outbreak of influenza A(H1N1) pandemic in 2009. Polymerase chain reaction (PCR) procedures established as part of a capacity-building process were used to conduct prospective respiratory surveillance in a region where few previous studies have been undertaken.
Methods: Between October 2008 and September 2010, nose and throat swabs from adults and children (approximately 20 per week) presenting with an acute respiratory illness to the Ninh Hoa General Hospital were collected. Same-day PCR testing and result reporting for 13 respiratory viruses were carried out by locally trained scientists.
Results: Of 2144 surveillance samples tested, 1235 (57.6%) were positive for at least one virus. The most common were influenza A strains (17.9%), with pandemic influenza A(H1N1) 2009 and seasonal H3N2 strain accounting for 52% and 43% of these, respectively. Other virus detections included: rhinovirus (12.4%), enterovirus (8.9%), influenza B (8.3%), adenovirus (5.3%), parainfluenza (4.7%), respiratory syncytial virus (RSV) (3.9%), human coronavirus (3.0%) and human metapneumovirus (0.3%). The detection rate was greatest in the 0–5 year age group. Viral co-infections were identified in 148 (6.9%) cases.
Discussion: The outbreak in 2009 of the influenza A(H1N1) pandemic strain provided a practical test of the laboratory’s pandemic plan. This study shows that the availability of appropriate equipment and molecular-based testing can contribute to important individual and public health outcomes in geographical locations susceptible to emerging infections.
Acute viral respiratory infections are an important cause of morbidity and hospitalization in Viet Nam where social and demographic conditions appear to heighten the risk of outbreaks capable of causing widespread disease and mortality. In Viet Nam, human infections with avian influenza A(H5N1) virus have occurred since 20031 and cases of severe acute respiratory syndrome (SARS) occurred in 2004. The socio-demographic and clinical features relating to these infections in Viet Nam have been previously described.2–7 Although SARS has not re-appeared, sporadic cases of human infection with avian influenza viruses continue to occur. As of November 2011, Viet Nam has recorded the third highest number of avian influenza cases and second highest number of related deaths globally.5 More recently, the rapid spread of the influenza A(H1N1) pandemic 2009 strain (hereafter referred to as A[H1N1]pdm09) into Viet Nam resulted in many thousands of laboratory-confirmed cases and 58 associated deaths during the first epidemic wave.8 Common non-influenza respiratory viruses are also important causes of significant acute respiratory infection in the country.9,10
The primary aim of this study was to assist the Virology Laboratory at the Nha Trang Pasteur Institute (NTPI) to develop laboratory preparedness for respiratory virus outbreaks, including the detection of common respiratory viruses and avian influenza viruses. Such laboratory capacity would provide early warning to Vietnamese public health authorities of an outbreak of infection of importance to human health, knowledge of which could be passed on to other countries in the region, including Australia. A second aim was to use the established polymerase chain reaction (PCR) methods to test respiratory samples collected from patients attending a local general hospital as proof of principal that staff training and transfer of technology were adequate. The surveillance period coincided with the occurrence of A(H1N1)pdm09, thereby enabling a practical assessment of the laboratory capacity that had been developed.
Specimens were collected from patients attending the Ninh Hoa General Hospital in Khanh Hoa province in rural south-central Viet Nam. The provincial capital city, Nha Trang, is located on the south-eastern coast, 38 km from the hospital. The region has a tropical climate with average temperatures ranging between 27°C and 33°C across two seasons: a dry season from January through August and a wet season from September until the end of December. The district population was 241 173 in 2009. In the same year, the 200-bed hospital treated a total of 19 516 patients, 927 with suspected pneumonia and 1654 with upper respiratory tract illnesses. The choice of the NTPI as the participating laboratory in Viet Nam was made by the Viet Nam Ministry of Health because laboratory capacity existed in the north (Ha Noi) and south (Ho Chi Minh City) but was less developed in the central regions of the country. Support to NTPI involved transfer of test methods and purchase of equipment suitable for rapid throughput testing. The established procedure required testing by locally trained Vietnamese scientists immediately on receipt of the samples, and same-day reporting.
Nose and throat swabs were collected from approximately 20 consenting patients each week between October 2008 and September 2010 inclusive. All patients had symptoms consistent with respiratory infection. Sample numbers were increased to approximately 40 per week during the outbreak of A(H1N1)pdm09. The swabs were pooled into 1.5ml of viral transport media, stored on-site at 4°C and then transported at 4°C twice weekly to NTPI where PCR testing for common respiratory viruses was performed on the day of arrival by local NTPI-employed scientists. During the 2009 outbreak, specimens not related to surveillance were also received at NTPI but were only tested for A(H1N1)pdm09. The presence of bacteria and other non-viral agents capable of causing respiratory symptoms was not sought in any samples. Surveillance samples were not collected during Vietnamese Lunar New Year occurring in January 2009 and February 2010.
Nucleic acid extraction and reverse transcriptionViral nucleic acid was extracted from 200µl of sample using a QIAxtractor robot (Qiagen, Valencia, CA, United States of America) and Qiagen DX reagent packs. The elution volume was 70µl. To control for the nucleic acid extraction, reverse transcription and PCR amplification steps, a non-human RNA virus (bovine viral diarrhoea virus [BVDV]) was spiked at low copy number into each sample before nucleic acid extraction and amplified using BVDV-specific primers. Reverse transcription was performed on 10µl of extract using random hexamer priming as previously described.11
Respiratory viruses were detected using multiplex PCR assays based on methods reported previously.11,12 The viruses detected were: influenza A, influenza B, parainfluenza virus (PIV) types 1, 2 and 3, respiratory syncytial virus (RSV), picornaviruses (rhinoviruses and enteroviruses), adenoviruses, human metapneumoviruses (hMPV), and human coronavirus (HCoV) types OC43, 229E and NL63. Modifications to the published methods involved replacement of subtype-specific (H1 and H3) primers with primers specific for the influenza A matrix gene and the addition of primers to enable HCoV-NL63 detection. The primer sequences targeting the influenza A virus matrix gene and HCoV-NL63 are shown in Table 1. Differentiation of rhinoviruses from enteroviruses involved testing of picornavirus-positive specimens in an enterovirus-specific nested PCR as follows: 2.5µl of cDNA was added to a final first-round mastermix volume of 40µl with primers ENTC and ENTD (Table 1). Subsequent cycling conditions were 95°C for three minutes followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 53°C and 30 seconds at 72°C, with final extension of five minutes at 72°C. Two microlitres of first-round PCR product were then transferred to a second round mastermix containing second-round primers ENTB and ENTC. Second round amplification was for 25 cycles at the conditions described above. The final PCR product was analysed on an agarose gel stained with ethidium bromide.
Subtyping of influenza A viruses detected before the emergence of A(H1N1)pdm09 was performed using a hemi-nested gel-based assay. During the 2009 outbreak, all samples were tested in two separate assays, a real-time PCR assay incorporating influenza A virus matrix and BVDV primers and the respiratory multiplex assay.11 A(H1N1)pdm09 was confirmed in influenza-positive samples using a real-time PCR assay incorporating specific primers. Samples testing negative for the pandemic strain were then tested for influenza A(H3N2) or seasonal influenza A(H1N1) as previously described.11 Identification of avian influenza H5N1 used a real-time PCR (primers and probe sequences shown in Table 1). The final reaction volume was 20µl with a thermal profile of 20 seconds at 95°C followed by 45 cycles of three seconds at 95°C and 30 seconds at 60°C.

A total of 2144 surveillance specimens collected from individual patients were tested, with one quarter of these cases admitted to a hospital ward (Table 2). During the A(H1N1)pdm09 outbreak in 2009, the laboratory also received 1541 specimens from patients attending hospitals other than Ninh Hoa for influenza virus testing only (these results are not included as part of the surveillance study). Surveillance patients ranged in age from 0.1 months to 85 years (mean 13 years; median seven years). The majority were children aged five years and under (45.1%). There were more males than females (55% versus 45% [Table 2]).
Of the 2144 surveillance samples tested, 1235 (57.6%) were positive for at least one virus (Table 2). The most common were influenza strains, more than half of which were A(H1N1)pdm09, first detected at the end of the dry season in 2009, then persisting through the wet season in that year (Table 2, Figure 1). This virus replaced an epidemic of A(H3N2) that occurred throughout the dry season in 2009. The A(H3N2) strain reappeared in 2010, peaking at the end of the dry season. A(H1N1)pdm09 infected mainly children and young adults, in contrast to influenza A(H3N2) infections, which mainly occurred in children under five-years-old (Table 3). Two large influenza B epidemics occurred during the study, peaking in the wet seasons of 2008 and 2010 but also circulating at low levels during the dry season in both years. Seasonal A(H1N1) strains were rarely detected. There was a single detection of non-pathogenic H5N1 in a three-year-old child (Table 3). This child had no known contact with poultry or other birds, but lived within one kilometre of an ostrich farm.
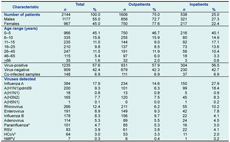
RSV - respiratory syncytial virus; HCoV - human coronavirus; and hMPV - human metapneumovirus.
* PIV-1 (53.5%), PIV-2 (13.9%), PIV-3 (26.7%), PIV-not typed (5.9%).
† HCoV-OC43 (72%), HCoV-229E (4.7%), HCoV-NL63 (23.4).
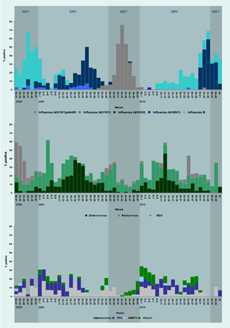
RSV - respiratory syncytial virus; PIV - parainfluenza virus; hMPV - human metapneumovirus; and HCoV - human coronavirus.

RSV - respiratory syncytial virus; hMPV - human metapneumovirus; and HCoV - human coronavirus.
Of the 1541 non-surveillance samples tested during June to December 2009 for the pandemic strain, 637 (41%) were positive.
Picornaviruses were detected throughout the two years of surveillance although only rhinoviruses circulated continuously (Table 2, Figure 1). Two large enterovirus outbreaks were mainly confined to the dry seasons in 2009 and 2010 (Figure 1). A similar age distribution was seen for both virus types (Table 3).
Several respiratory viruses were detected only rarely, including adenovirus (5.3% of the total number of viruses detected), PIV (4.7%), RSV (3.9%), HCoV (3%) and hMPV (0.3%). Parainfluenza viruses, of which type 1 was the most common, circulated throughout both 2009 and 2010 (Table 2). RSV infections, which were not associated with an obvious seasonal distribution, occurred in the very young, adults of childbearing age and the elderly. HCoVs were also detected throughout the year across all age groups, albeit in low numbers. OC43 was the most common coronavirus, with a distribution that spanned the wet and dry seasons in 2008 through 2009 and 2009 through 2010 (Table 2, Figure 1).
There were 148 cases (6.9%) involving co-infection with at least two viruses (Table 4). Rhinoviruses (50.7% of all co-infections) and adenoviruses (39.9%) were most commonly involved.
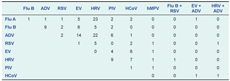
Flu A - influenza virus A; Flu B - influenza virus B; ADV - adenovirus; RSV - respiratory syncytial virus; EV - enterovirus; HRV - human rhinovirus; PIV - parainfluenza virus; HCoV - human coronavirus; and hMPV - human metapneumovirus.
The major aim of this study was to build permanent laboratory capacity that could provide early warning to public health authorities of an emerging epidemic of highly pathogenic avian influenza virus or other respiratory viruses. Over the two-year study period, 2144 samples were tested by locally trained Vietnamese scientists, with over half being positive for at least one virus. Viruses detected included influenza, most commonly A(H1N1)pdm09, as well as rhinovirus, enterovirus and adenovirus. The outbreak in 2009 of the influenza A(H1N1) pandemic strain provided an important opportunity for the laboratory to function under pandemic conditions, with an additional 1541 specimens received.
Two other studies of respiratory viruses have recently been undertaken in Viet Nam. Both involved hospitalized children living in either the central region where we conducted our investigation or in south Viet Nam.9,10 The study in the south was carried out over more than three years between 2004 and 2008, enabling the seasonal distribution of respiratory viruses to be investigated. It revealed a similar pattern of circulation of influenza to that which we subsequently observed, with peaks occurring in the wet seasons and lower levels of circulation at other times. Although the study in south Viet Nam did not distinguish between the circulation of influenza A and B, our study showed that both virus types could be detected throughout the year and that conditions prevailing in the wet season favoured increased levels of circulation and infection.
The transmission of A(H1N1)pdm09 in central Viet Nam shared similarities and differences from that experienced in other geographical locations. In 2009 in Viet Nam, an established outbreak of A(H3N2) was quickly replaced by the pandemic virus, as observed in temperate climates including Australia, the United States of America and Europe.13–15 However, in contrast to the dominance of A(H1N1)pdm09 during subsequent winter influenza seasons in Australia in 2010 and Europe in 2010 and 2011,16,17 the pandemic virus only returned in very low numbers in and around Nha Trang in 2010. In the first post-pandemic influenza season in 2010, co-circulation of A(H3N2) and influenza type B was observed in this area of Viet Nam, more closely resembling influenza activity patterns observed in northern Asia, the United States of America and Canada.18
RSV and hMPV circulated only in the wet season in central Viet Nam, consistent with the previous study in the south of the country.10 In contrast, coronaviruses, rhinoviruses and adenoviruses circulated throughout the year. The adenoviruses were often associated with co-infections, making their clinical significance unclear. Parainfluenza virus circulation spanned wet and dry seasons.
Enteroviruses causing neurological disease,19,20 hand, foot and mouth disease and gastroenteritis have been reported in Viet Nam,19,21 but their association with respiratory syndromes has not been investigated. In our study, enterovirus circulation was restricted to the dry season in both study years. Identification of the enterovirus serotypes involved is currently being undertaken and is of interest since not all serotypes are thought to cause respiratory symptoms. A previous study involving hospitalized children in south Viet Nam also demonstrated the important role of enteroviruses in respiratory disease, but unlike our study did not fully reveal the role of rhinoviruses, probably because testing for type C rhinoviruses was not undertaken.10 In contrast, previous surveillance in Nha Trang revealed a significant number of cases attributable to rhinovirus infection but did not include testing that would most likely have indicated the importance of enteroviruses.9
A limitation of our study was that it was not primarily a clinical investigation and therefore we did not have access to detailed clinical information on the patients from whom specimens were collected. Hence, we can only make limited conclusions regarding the clinical impact of the viruses detected.
Overall only one quarter of patients from whom samples were obtained were admitted to a hospital ward. The exception was during the A(H1N1)pdm09 outbreak when more than half of the laboratory-confirmed cases were admitted. Following the first appearance of the pandemic strain in central Viet Nam, suspected cases appear to have been admitted as a precaution while information on the clinical severity of this novel strain was gathered over time.
Because our study involved both children and adults, in contrast to the two previously reported molecular-based studies in Viet Nam involving only children,9,10 we were able to show that the circulation and morbidity associated with common viruses such as influenza and less common viruses such as RSV, PIV and coronaviruses, had a wide age demographic. To some extent the intervention of A(H1N1)pdm09 hampered our efforts to comprehensively study the seasonality of respiratory viruses in this region of Viet Nam because the scale of the outbreak minimized the likelihood of detecting other circulating respiratory viruses. However, it did provide the opportunity for an assessment of a pandemic plan that had only been established in the previous year. The detection of an influenza H5 virus, albeit a non-pathogenic strain, in a patient presenting with symptoms not suggestive of influenza infection, prompted an investigation of asymptomatic household contacts, each of whom returned negative results. During the 2009 influenza outbreak, locally trained medical, nursing and scientific personnel collected, transported and tested large numbers of specimens within a short turnaround time. Transfer of technology and the training of local scientists were the essential elements in this process. Our study shows that the availability of appropriate equipment and molecular-based diagnostic testing contributes to important individual and public health outcomes in geographical locations susceptible to emerging infections.
None declared.
This study was conceived and funded by the Australian Agency for International Development under its Australia-Viet Nam Laboratory Partnership Programme.